FieldTrip beamforming 2
Contents
- Get data and choose time of interest
- Prepare head model and MRI (from template)
- FieldTrip version of Beamforming
- Compute cov for averaged data, toi only
- Compute cov for the whole averaged data
- Same (whole averaged) but with LCMV
- Compute cov for raw data
- Robinson's SAM in FieldTrip envelope
- MNE
- compute MNE and display time of interest
- make MNE movie
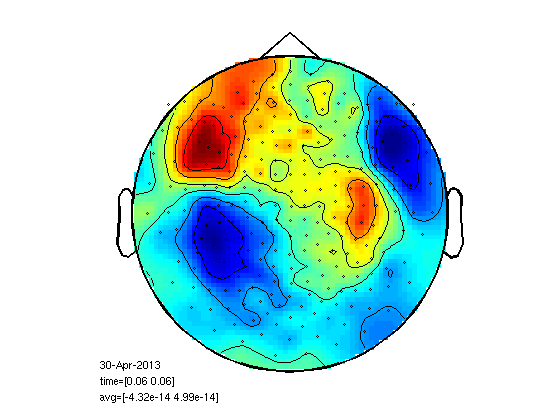
Get data and choose time of interest
% first go back to course8 and make sure you already run SAMwts with -t % pnt.txt option. cd oddball load trl % first we need data source='c,rfhp0.1Hz'; cfg=[]; cfg.dataset=source; cfg.trialfun='trialfun_beg'; cfg2=ft_definetrial(cfg); cfg2.trl=trl; cfg2.demean='yes'; cfg2.baselinewindow=[-0.2 0]; cfg2.bpfilter='yes'; cfg2.bpfreq=[3 30]; cfg2.channel='MEG'; data=ft_preprocessing(cfg2); cfg=[]; cfg.trials=find(data.trialinfo==128); standard=ft_preprocessing(cfg,data); cfg=[]; cfg.method='summary'; cfg.alim=1e-12; standard=ft_rejectvisual(cfg, standard); stdAvg=ft_timelockanalysis([],standard); cfg=[]; cfg.xlim=[0.06 0.06]; cfg.layout='4D248.lay'; cfg.channel='MEG'; ft_topoplotER(cfg,stdAvg); toi=[0.043657 0.075163];
evaluating trialfunction 'trialfun_beg'
reading header...
applying digital weights in the gradiometer balancing matrix
undoing the Supine balancing
found 1 events
created 1 trials
the call to "ft_definetrial" took 1 seconds and an estimated 0 MB
applying digital weights in the gradiometer balancing matrix
undoing the Supine balancing
processing channel { 'A22' 'A2' 'A104' 'A241' 'A138' 'A214' 'A71' 'A26' 'A93' 'A39' 'A125' 'A20' 'A65' 'A9'
reading and preprocessing
reading and preprocessing trial 1 from 347
reading and preprocessing trial 2 from 347
reading and preprocessing trial 3 from 347
the call to "ft_preprocessing" took 16 seconds and an estimated 580 MB
selecting 239 trials
preprocessing
preprocessing trial 1 from 239
preprocessing trial 2 from 239
.
.
.
the call to "ft_preprocessing" took 0 seconds and an estimated 2 MB
the input is raw data with 248 channels and 239 trials
showing a summary of the data for all channels and trials
computing metric [---------------------------------------------------------]
223 trials marked as GOOD, 16 trials marked as BAD
248 channels marked as GOOD, 0 channels marked as BAD
the following trials were removed: 5, 6, 16, 30, 34, 45, 56, 86, 96, 105, 129, 133, 164, 199, 200, 236
the call to "ft_rejectvisual" took 14 seconds and an estimated 89 MB
the input is raw data with 248 channels and 223 trials
averaging trials
averaging trial 1 of 223
averaging trial 2 of 223
averaging trial 3 of 223
.
.
.
the call to "ft_timelockanalysis" took 0 seconds and an estimated 0 MB
reading layout from file 4D248.lay
the call to "ft_prepare_layout" took 0 seconds and an estimated 0 MB
the call to "ft_topoplotTFR" took 0 seconds and an estimated 0 MB
the call to "ft_topoplotER" took 0 seconds and an estimated 0 MB
Prepare head model and MRI (from template)
if exist('headmodel.mat','file') load headmodel % it was created in course8 (and 7) else [vol,grid,mesh,M1,single]=headmodel_BIU([],[],[],[],'localspheres'); end load ~/ft_BIU/matlab/files/sMRI.mat mri_realign=sMRI; mri_realign.transform=inv(M1)*sMRI.transform;
FieldTrip version of Beamforming
Compute cov for averaged data, toi only
compute cov for very short data, time of interest and baseline
source=OBbeamform(stdAvg,toi,'sam',mri_realign)
converting units from 'm' to 'mm'
the input is timelock data with 248 channels and 1018 timebins
Warning: the trial definition in the configuration is inconsistent with the actual data
Warning: reconstructing sampleinfo by assuming that the trials are consecutive segments of a
continuous recording
averaging trials
averaging trial 1 of 1
.
.
.
the call to "ft_timelockanalysis" took 0 seconds and an estimated 0 MB
the input is timelock data with 248 channels and 1018 timebins
averaging trials
averaging trial 1 of 1
.
.
.
the call to "ft_timelockanalysis" took 0 seconds and an estimated 0 MB
the input is timelock data with 248 channels and 1018 timebins
Warning: The field cfg.lambda is deprecated, please specify it as cfg.sam.lambda instead of cfg.
using headmodel specified in the configuration
using gradiometers specified in the data
creating dipole grid based on user specified dipole positions
3480 dipoles inside, 795 dipoles outside brain
the call to "ft_prepare_sourcemodel" took 0 seconds and an estimated 0 MB
scanning repetition 1
Warning: covariance matrix is rank deficient
scanning grid
Warning: The input units are unknown for points and S/unknown for conductivity
scanning grid 1/3480
scanning grid 2/3480
scanning grid 3/3480
.
.
.
scanning grid 3478/3480
scanning grid 3479/3480
scanning grid 3480/3480
the call to "ft_sourceanalysis" took 72 seconds and an estimated 0 MB
the input is timelock data with 248 channels and 1018 timebins
Warning: The field cfg.lambda is deprecated, please specify it as cfg.sam.lambda instead of cfg.
using headmodel specified in the configuration
using gradiometers specified in the data
creating dipole grid based on user specified dipole positions
3480 dipoles inside, 795 dipoles outside brain
the call to "ft_prepare_sourcemodel" took 0 seconds and an estimated 0 MB
scanning repetition 1
Warning: covariance matrix is rank deficient
scanning grid 1/3480
scanning grid 2/3480
scanning grid 3/3480
.
.
.
scanning grid 3478/3480
scanning grid 3479/3480
scanning grid 3480/3480
the call to "ft_sourceanalysis" took 72 seconds and an estimated 0 MB
Warning: SOURCEINTERPOLATE is only a compatibility wrapper, which will soon be removed. Please
instead call FT_SOURCEINTERPOLATE.
the input is source data with 4275 positions
the input is volume data with dimensions [91 109 91]
selecting subvolume of 48.8%
interpolating
interpolating 0.0%
interpolating 5.0%
interpolating 95.0%
interpolating 100.0%
reslicing and interpolating avg.nai
interpolating
interpolating 0.0%
interpolating 5.0%
interpolating 95.0%
interpolating 100.0%
the call to "ft_sourceinterpolate" took 1 seconds and an estimated 0 MB
the input is volume data with dimensions [91 109 91]
not downsampling anatomy
not downsampling avg.nai
not downsampling inside
the call to "ft_volumedownsample" took 0 seconds and an estimated 0 MB
no masking parameter
click with mouse button to reposition the cursor
press n/l/r on keyboard to record a fiducial position
press q on keyboard to quit interactive mode
voxel 361680, indices [46 51 37], location [5.8 2.1 48.9] mm, value 218.015819
scaling anatomy
scaling anatomy
scaling anatomy
nas = undefined
lpa = undefined
rpa = undefined
click with mouse button to reposition the cursor
press n/l/r on keyboard to record a fiducial position
press q on keyboard to quit interactive mode
voxel 361802, indices [77 52 37], location [8.2 53.0 48.8] mm, value 71.405395
scaling anatomy
scaling anatomy
scaling anatomy
nas = undefined
lpa = undefined
rpa = undefined
click with mouse button to reposition the cursor
press n/l/r on keyboard to record a fiducial position
press q on keyboard to quit interactive mode
voxel 351792, indices [77 51 36], location [6.1 53.0 47.9] mm, value 74.522614
scaling anatomy
scaling anatomy
scaling anatomy
the call to "ft_sourceplot" took 645 seconds and an estimated 1 MB
source =
dim: [15 19 15]
time: [1x1018 double]
pos: [4275x3 double]
inside: [3480x1 double]
outside: [795x1 double]
method: 'average'
avg: [1x1 struct]
cfg: [1x1 struct]
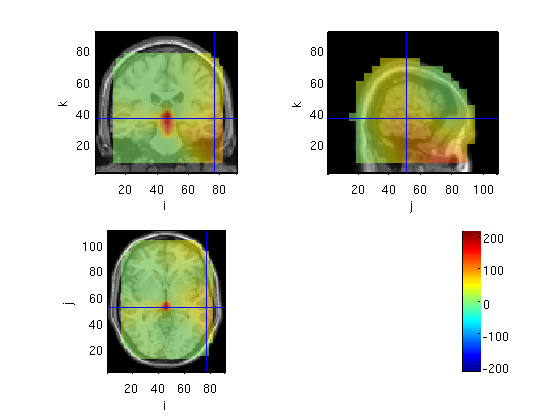
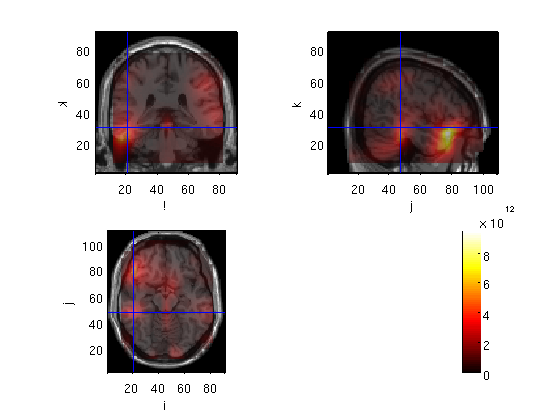
Compute cov for the whole averaged data
source=OBbeamform1(stdAvg,toi,'sam',mri_realign)
converting units from 'm' to 'mm'
Warning: TIMELOCKANALYSIS is only a compatibility wrapper, which will soon be removed. Please
instead call FT_TIMELOCKANALYSIS.
the input is timelock data with 248 channels and 1018 timebins
Warning: the trial definition in the configuration is inconsistent with the actual data
Warning: reconstructing sampleinfo by assuming that the trials are consecutive segments of a
continuous recording
Warning: the option cfg.covariancewindow is not specified, taking all time points
averaging trials
averaging trial 1 of 1
the call to "ft_timelockanalysis" took 0 seconds and an estimated 0 MB
the input is timelock data with 248 channels and 1018 timebins
Warning: The field cfg.lambda is deprecated, please specify it as cfg.sam.lambda instead of cfg.
using headmodel specified in the configuration
using gradiometers specified in the data
creating dipole grid based on user specified dipole positions
3480 dipoles inside, 795 dipoles outside brain
the call to "ft_prepare_sourcemodel" took 0 seconds and an estimated 0 MB
scanning repetition 1
Warning: covariance matrix is rank deficient
scanning grid
Warning: The input units are unknown for points and S/unknown for conductivity
scanning grid 1/3480
scanning grid 2/3480
scanning grid 3/3480
.
.
.
scanning grid 3479/3480
scanning grid 3480/3480
the call to "ft_sourceanalysis" took 73 seconds and an estimated 0 MB
Warning: SOURCEINTERPOLATE is only a compatibility wrapper, which will soon be removed. Please
instead call FT_SOURCEINTERPOLATE.
the input is source data with 4275 positions
the input is volume data with dimensions [91 109 91]
selecting subvolume of 48.8%
interpolating
interpolating 0.0%
interpolating 5.0%
interpolating 95.0%
interpolating 100.0%
reslicing and interpolating avg.nai
interpolating
interpolating 0.0%
interpolating 5.0%
interpolating 90.0%
interpolating 95.0%
interpolating 100.0%
the call to "ft_sourceinterpolate" took 1 seconds and an estimated 0 MB
the input is volume data with dimensions [91 109 91]
not downsampling anatomy
not downsampling avg.nai
not downsampling inside
the call to "ft_volumedownsample" took 0 seconds and an estimated 0 MB
no masking parameter
click with mouse button to reposition the cursor
press n/l/r on keyboard to record a fiducial position
press q on keyboard to quit interactive mode
voxel 610691, indices [81 62 62], location [35.4 59.6 79.0] mm, value 1229.834276
scaling anatomy
scaling anatomy
scaling anatomy
the call to "ft_sourceplot" took 68 seconds and an estimated 0 MB
source =
dim: [15 19 15]
time: [1x1018 double]
pos: [4275x3 double]
inside: [3480x1 double]
outside: [795x1 double]
method: 'average'
avg: [1x1 struct]
cfg: [1x1 struct]
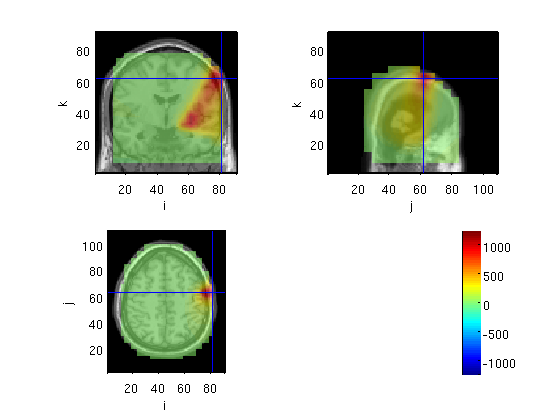
Same (whole averaged) but with LCMV
source=OBbeamform1(stdAvg,toi,'lcmv',mri_realign)
converting units from 'm' to 'mm'
Warning: TIMELOCKANALYSIS is only a compatibility wrapper, which will soon be removed. Please
instead call FT_TIMELOCKANALYSIS.
the input is timelock data with 248 channels and 1018 timebins
Warning: the trial definition in the configuration is inconsistent with the actual data
Warning: reconstructing sampleinfo by assuming that the trials are consecutive segments of a
continuous recording
Warning: the option cfg.covariancewindow is not specified, taking all time points
averaging trials
averaging trial 1 of 1
the call to "ft_timelockanalysis" took 0 seconds and an estimated 0 MB
the input is timelock data with 248 channels and 1018 timebins
Warning: The field cfg.keepfilter is deprecated, please specify it as cfg.lcmv.keepfilter instead
of cfg.
Warning: The field cfg.lambda is deprecated, please specify it as cfg.lcmv.lambda instead of cfg.
using headmodel specified in the configuration
using gradiometers specified in the data
creating dipole grid based on user specified dipole positions
3480 dipoles inside, 795 dipoles outside brain
the call to "ft_prepare_sourcemodel" took 0 seconds and an estimated 0 MB
scanning repetition 1
Warning: covariance matrix is rank deficient
scanning grid
Warning: The input units are unknown for points and S/unknown for conductivity
scanning grid 1/3480
scanning grid 2/3480
scanning grid 3/3480
.
.
.
scanning grid 3478/3480
scanning grid 3479/3480
scanning grid 3480/3480
the call to "ft_sourceanalysis" took 24 seconds and an estimated 0 MB
Warning: SOURCEINTERPOLATE is only a compatibility wrapper, which will soon be removed. Please
instead call FT_SOURCEINTERPOLATE.
the input is source data with 4275 positions
the input is volume data with dimensions [91 109 91]
selecting subvolume of 48.8%
interpolating
interpolating 0.0%
interpolating 95.0%
interpolating 100.0%
reslicing and interpolating avg.nai
interpolating
interpolating 0.0%
interpolating 5.0%
interpolating 95.0%
interpolating 100.0%
the call to "ft_sourceinterpolate" took 1 seconds and an estimated 0 MB
the input is volume data with dimensions [91 109 91]
not downsampling anatomy
not downsampling avg.nai
not downsampling inside
the call to "ft_volumedownsample" took 0 seconds and an estimated 0 MB
no masking parameter
click with mouse button to reposition the cursor
press n/l/r on keyboard to record a fiducial position
press q on keyboard to quit interactive mode
voxel 115641, indices [71 72 12], location [33.9 43.0 4.6] mm, value 98.441667
scaling anatomy
scaling anatomy
scaling anatomy
the call to "ft_sourceplot" took 6 seconds and an estimated 0 MB
source =
dim: [15 19 15]
time: [1x1018 double]
pos: [4275x3 double]
inside: [3480x1 double]
outside: [795x1 double]
method: 'average'
avg: [1x1 struct]
cfg: [1x1 struct]
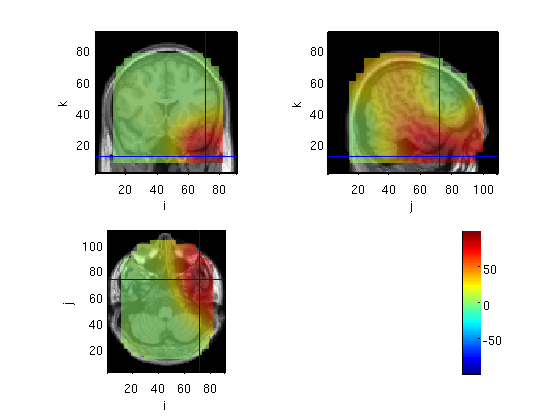
Compute cov for raw data
source=OBbeamform(standard,toi,'sam',mri_realign)
converting units from 'm' to 'mm'
Warning: TIMELOCKANALYSIS is only a compatibility wrapper, which will soon be removed. Please
instead call FT_TIMELOCKANALYSIS.
the input is raw data with 248 channels and 223 trials
selecting 100 trials
selecting 100 trials
Warning: the option cfg.covariancewindow is not specified, taking all time points
averaging trials
averaging trial 1 of 100
averaging trial 2 of 100
averaging trial 3 of 100
.
.
.
averaging trial 100 of 100
the call to "ft_timelockanalysis" took 1 seconds and an estimated 193 MB
the input is timelock data with 248 channels and 1018 timebins
Warning: The field cfg.lambda is deprecated, please specify it as cfg.sam.lambda instead of cfg.
using headmodel specified in the configuration
using gradiometers specified in the data
creating dipole grid based on user specified dipole positions
3480 dipoles inside, 795 dipoles outside brain
the call to "ft_prepare_sourcemodel" took 0 seconds and an estimated 0 MB
scanning repetition 1
scanning grid
scanning grid 1/3480
scanning grid 2/3480
.
.
.
scanning grid 3478/3480
scanning grid 3479/3480
scanning grid 3480/3480
the call to "ft_sourceanalysis" took 72 seconds and an estimated 193 MB
Warning: TIMELOCKANALYSIS is only a compatibility wrapper, which will soon be removed. Please
instead call FT_TIMELOCKANALYSIS.
the input is raw data with 248 channels and 223 trials
averaging trials
averaging trial 1 of 223
averaging trial 2 of 223
averaging trial 3 of 223
.
.
.
averaging trial 222 of 223
averaging trial 223 of 223
the call to "ft_timelockanalysis" took 0 seconds and an estimated 193 MB
Warning: SOURCEINTERPOLATE is only a compatibility wrapper, which will soon be removed. Please
instead call FT_SOURCEINTERPOLATE.
the input is source data with 4275 positions
the input is volume data with dimensions [91 109 91]
selecting subvolume of 48.8%
interpolating
interpolating 0.0%
interpolating 5.0%
interpolating 95.0%
interpolating 100.0%
reslicing and interpolating avg.nai
interpolating
interpolating 0.0%
interpolating 5.0%
interpolating 95.0%
interpolating 100.0%
the call to "ft_sourceinterpolate" took 1 seconds and an estimated 0 MB
the input is volume data with dimensions [91 109 91]
not downsampling anatomy
not downsampling avg.nai
not downsampling inside
the call to "ft_volumedownsample" took 0 seconds and an estimated 0 MB
no masking parameter
click with mouse button to reposition the cursor
press n/l/r on keyboard to record a fiducial position
press q on keyboard to quit interactive mode
voxel 362610, indices [66 61 37], location [23.9 34.9 44.5] mm, value 1183.535679
scaling anatomy
scaling anatomy
scaling anatomy
the call to "ft_sourceplot" took 4 seconds and an estimated 0 MB
source =
dim: [15 19 15]
time: [1x1018 double]
pos: [4275x3 double]
inside: [3480x1 double]
outside: [795x1 double]
method: 'average'
avg: [1x1 struct]
cfg: [1x1 struct]
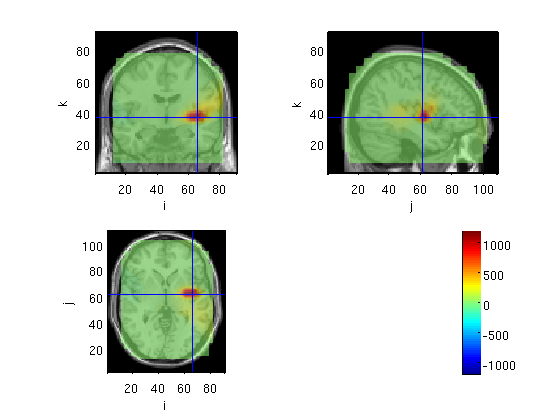
Robinson's SAM in FieldTrip envelope
here we use the output of SAMwts as filter similar to cov for raw data
load('SAM/pnt.txt.mat') filter=wts2filter(ActWgts,grid.inside,size(grid.outside,1)); source=OBbeamform(stdAvg,toi,'SAM',mri_realign,filter)
converting units from 'm' to 'mm'
Warning: SOURCEINTERPOLATE is only a compatibility wrapper, which will soon be removed. Please
instead call FT_SOURCEINTERPOLATE.
the input is source data with 4275 positions
the input is volume data with dimensions [91 109 91]
selecting subvolume of 48.8%
interpolating
interpolating 0.0%
interpolating 5.0%
interpolating 95.0%
interpolating 100.0%
reslicing and interpolating avg.nai
interpolating
interpolating 0.0%
interpolating 5.0%
interpolating 95.0%
interpolating 100.0%
the call to "ft_sourceinterpolate" took 1 seconds and an estimated 0 MB
the input is volume data with dimensions [91 109 91]
not downsampling anatomy
not downsampling avg.nai
not downsampling inside
the call to "ft_volumedownsample" took 0 seconds and an estimated 0 MB
no masking parameter
click with mouse button to reposition the cursor
press n/l/r on keyboard to record a fiducial position
press q on keyboard to quit interactive mode
voxel 212020, indices [81 41 22], location [-16.8 59.4 33.2] mm, value 358.764443
scaling anatomy
scaling anatomy
scaling anatomy
the call to "ft_sourceplot" took 2 seconds and an estimated 0 MB
source =
pos: [4275x3 double]
inside: [3480x1 double]
outside: [795x1 double]
dim: [15 19 15]
avg: [1x1 struct]
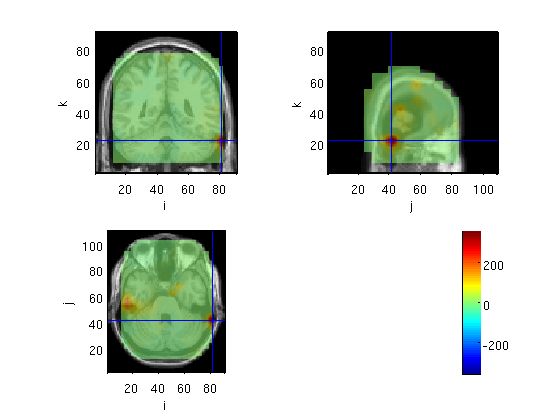
MNE
source=OBmne(stdAvg,toi,mri_realign)
converting units from 'm' to 'mm'
Warning: TIMELOCKANALYSIS is only a compatibility wrapper, which will soon be removed. Please
instead call FT_TIMELOCKANALYSIS.
the input is timelock data with 248 channels and 1018 timebins
Warning: the trial definition in the configuration is inconsistent with the actual data
Warning: reconstructing sampleinfo by assuming that the trials are consecutive segments of a
continuous recording
Warning: the option cfg.covariancewindow is not specified, taking all time points
averaging trials
averaging trial 1 of 1
the call to "ft_timelockanalysis" took 0 seconds and an estimated 0 MB
the input is timelock data with 248 channels and 1018 timebins
Warning: The field cfg.lambda is deprecated, please specify it as cfg.mne.lambda instead of cfg.
Warning: The field cfg.keepfilter is deprecated, please specify it as cfg.mne.keepfilter instead of
cfg.
using headmodel specified in the configuration
using gradiometers specified in the data
creating dipole grid based on user specified dipole positions
3480 dipoles inside, 795 dipoles outside brain
the call to "ft_prepare_sourcemodel" took 0 seconds and an estimated 0 MB
estimating current density distribution for repetition 1
computing forward model
Warning: The input units are unknown for points and S/unknown for conductivity
computing MNE source reconstruction, this may take some time...
computing the solution where the noise covariance is used for regularisation
the call to "ft_sourceanalysis" took 22 seconds and an estimated 1 MB
Warning: SOURCEINTERPOLATE is only a compatibility wrapper, which will soon be removed. Please
instead call FT_SOURCEINTERPOLATE.
the input is source data with 4275 positions
the input is volume data with dimensions [91 109 91]
selecting subvolume of 48.8%
interpolating
interpolating 0.0%
interpolating 5.0%
interpolating 95.0%
interpolating 100.0%
reslicing and interpolating avg.nai
interpolating
interpolating 0.0%
interpolating 5.0%
interpolating 95.0%
interpolating 100.0%
the call to "ft_sourceinterpolate" took 1 seconds and an estimated 0 MB
the input is volume data with dimensions [91 109 91]
not downsampling anatomy
not downsampling avg.nai
not downsampling inside
the call to "ft_volumedownsample" took 0 seconds and an estimated 0 MB
no masking parameter
click with mouse button to reposition the cursor
press n/l/r on keyboard to record a fiducial position
press q on keyboard to quit interactive mode
voxel 506921, indices [51 12 52], location [-57.2 10.4 88.2] mm, value 7.081434
scaling anatomy
scaling anatomy
scaling anatomy
the call to "ft_sourceplot" took 5 seconds and an estimated 0 MB
source =
dim: [15 19 15]
time: [1x1018 double]
pos: [4275x3 double]
inside: [1x3480 double]
outside: [1x795 double]
method: 'average'
avg: [1x1 struct]
cfg: [1x1 struct]
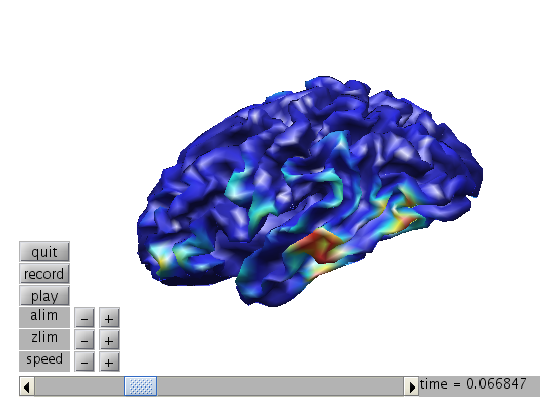
Compute MNE and display time of interest.
cfg = []; cfg.covariance = 'yes'; cfg.covariancewindow = [-inf 0]; cov = ft_timelockanalysis(cfg, stdAvg); [vol,grid,mesh,M1]=headmodel_BIU([],[],5,[],'localspheres'); cfg = []; cfg.channel ='MEG'; cfg.grid.pos = mesh.tess_ctx.vert; cfg.grid.inside = [1:size(mesh.tess_ctx.vert,1)]'; cfg.vol = vol; cfg.grad=ft_convert_units(stdAvg.grad,'mm'); leadfield = ft_prepare_leadfield(cfg); cfg=[]; cfg.method = 'mne'; cfg.grid = leadfield; cfg.vol = vol; cfg.mne.lambda = 1.2e9; %cfg.mne.lambda = 1e9; cov.grad=ft_convert_units(cov.grad,'mm') source = ft_sourceanalysis(cfg,cov); figure; bnd.pnt=mesh.tess_ctx.vert; bnd.tri=mesh.tess_ctx.face; ft_plot_mesh(bnd, 'vertexcolor', source.avg.pow(:,264));
the input is timelock data with 248 channels and 1018 timebins
Warning: the trial definition in the configuration is inconsistent with the actual data
Warning: reconstructing sampleinfo by assuming that the trials are consecutive segments of a
continuous recording
averaging trials
averaging trial 1 of 1
the call to "ft_timelockanalysis" took 0 seconds and an estimated 0 MB
evaluating trialfunction 'trialfun_beg'
reading header...
applying digital weights in the gradiometer balancing matrix
undoing the Supine balancing
found 1 events
created 1 trials
the call to "ft_definetrial" took 1 seconds and an estimated 0 MB
applying digital weights in the gradiometer balancing matrix
undoing the Supine balancing
processing channel { 'TRIGGER' 'RESPONSE' 'MLzA' 'MLyA' 'MLzaA' 'MLyaA' 'MLxA' 'A22' 'MLxaA' 'A2' 'MRzA' 'MRxA'
reading and preprocessing
reading and preprocessing trial 1 from 1
the call to "ft_preprocessing" took 1 seconds and an estimated 0 MB
channel = GyxA, not enough surface points, using all points
the call to "ft_prepare_localspheres_mm" took 5 seconds and an estimated 0 MB
undoing the Supine balancing
single sphere, 2562 surface points, center = [11.7 1.9 45.2], radius = 80.7
creating dipole grid based on user specified 3D grid
26381 dipoles inside, 4736 dipoles outside brain
converting units from 'm' to 'mm'
using headmodel specified in the configuration
using gradiometers specified in the configuration
creating dipole grid based on user specified dipole positions
8196 dipoles inside, 0 dipoles outside brain
the call to "ft_prepare_sourcemodel" took 0 seconds and an estimated 0 MB
computing leadfield
computing leadfield 1/8196
Warning: The input units are unknown for points and S/unknown for conductivity
computing leadfield 2/8196
computing leadfield 3/8196
.
.
.
computing leadfield 8195/8196
computing leadfield 8196/8196
the call to "ft_prepare_leadfield" took 52 seconds and an estimated 0 MB
converting units from 'm' to 'mm'
cov =
avg: [248x1018 double]
var: [248x1018 double]
time: [1x1018 double]
dof: [248x1018 double]
label: {248x1 cell}
dimord: 'chan_time'
cov: [248x248 double]
grad: [1x1 struct]
cfg: [1x1 struct]
the input is timelock data with 248 channels and 1018 timebins
using headmodel specified in the configuration
using gradiometers specified in the data
creating dipole grid based on user specified dipole positions
8196 dipoles inside, 0 dipoles outside brain
the call to "ft_prepare_sourcemodel" took 0 seconds and an estimated 0 MB
estimating current density distribution for repetition 1
using specified forward model
computing MNE source reconstruction, this may take some time...
computing the solution where the noise covariance is used for regularisation
taking pseudo-inverse due to large condition number
the call to "ft_sourceanalysis" took 2 seconds and an estimated 64 MB
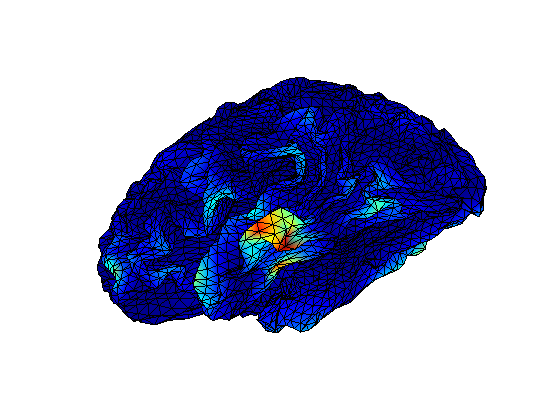
make MNE movie
source.tri=bnd.tri;
ft_sourcemovie([],source);
% you can alternatively use freesurfer and Harvard MNE software
the input is source data with 3000 positions baseline correcting dipole moments [--------------------------------------/] projecting dipole moment [------------------------------------------------/] computing power [---------------------------------------------------------/] the call to "ft_sourcedescriptives" took 6 seconds and an estimated 0 MB the input is source data with 3000 positions